We study the cell biology of bacteria, whose awesome experimental power allows us to attack questions with an integrated combination of genetics, biochemistry, microscopy, genomics, quantitative analysis, and computation. Our interdisciplary efforts are aided by our wonderful group of Princeton friends and collaborators.
We work on a wide range of systems with the unified theme of undersanding both the mechanisms and functions of self-assembly and cellular interactions. We are currently focusing on three inter-related areas:
Bacterial Self-Organization: Growth and Cell Shape
How do organisms like bacteria make such amazing and complex structures out of such simple building blocks? Our lab has discovered fundamental mechanisms by which self-assembly happens across a wide range of spatial scales, from atoms and metabolites to proteins and polymers to cells and communities. For example, by assembling nanometer-sized proteins into micron-sized cytoskeletal polymers, bacteria make the measurements necessary to achieve their characteristic shapes. And by modulating ribosomes in different ways bacteria can achieve similar growth rates in different nutrient conditions. We are now working on the mechanisms by which self-assembly occurs as well as its functions in cell shape, metabolism, and pathogenesis. We are also investigating growth from both surface area and biomass perspectives.
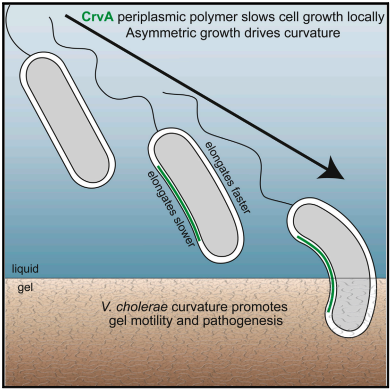
Spheres, rods, commas, helices, branches, cubes, stars are just some of the tremendous diversity of cell shapes found in the bacterial kingdom. To generate such a wide array of morphologies in a single cell, bacteria must organize growth in a tiny ~1 μm3 space. But how? Work from our lab and others has shown that the proteins of the bacterial cytoskeleton self-assemble into cellular-scale structures that play essential roles in virtually all cell biological processes. We are currently studying what proteins constitute the bacterial cytoskeleton, how cytoskeletal proteins assemble, and how they regulate processes such as cell shape, division, polarity, chromosome dynamics, metabolism, and pathogenesis. To understand both species-specific and generalizable features we perform our studies in a wide range of bacterial species including the polarized bacterium Caulobacter crescentus, whose asymmetric shape and synchronizable cell cycle make it particularly attractive for cell biological studies, the incredibly-well-characterized bacterium E. coli, and human pathogens like Pseudomonas aeruginosa and Vibrio cholerae. These directions also enable us to study the selective benefits that specific shapes and polymers confer to each species, letting us ask not only how each shape is generated, but why.
Microbe-host interactions: the cell biology of pathogenesis and environmental sensing
Once built, how do those bacteria interact with each other, their hosts, and their environments? Our lab discovered that pathogens like P. aeruginosa use mechanosensing to sense the presence of host cells and induce virulence pathways. Meanwhile, hosts can sense the presence of specific bacterial species based on their sRNA sequences. We are now focusing on novel mechanisms and functions of these and other forms of microbe-host interactions.
We can now use microfluidics and other methods to engineer environments that better mimic the mechanical, chemical, and biological cues that bacteria naturally encounter. We found that the polar adhesion and motility structures of species such as Caulobacter crescentus and Pseudomonas aeruginosa are essential features that shape their colonization strategies. We are discovered that bacteria can sense and respond to their mechanical environments. We are currently studying how bacterial mechanosensation occurs and the role of bacterial organization in shaping bacterial interactions with their hosts and with other bacteria. We are also interested in how hosts sense the presence of bacteria to discriminate benficial and harmful species, and how bacteria and hosts coexist in stable microbiome communities.
Antibiotics: Quantitative approaches to discovering and understanding new drugs for superbugs
How can we use our quantitative tools to both discover and characterize novel antibiotics? The rise of antibiotic resistance has made the discovery of novel antibiotics an urgent societal threat, yet the pipeline of truly novel antibiotics has largely dried up. We have used a combination of imaging, machine learning, and systems approaches to develop new approaches to rapidly determining antibiotic mechanisms of action. We used these methods to discover one of the first novel antibiotics to be found in decades. We are now focusing on understanding how these novel antibiotics function, discovering additional novel antibiotics, and using antibiotics to probe previously unappreciated features of bacterial cell biology. We are also working on ways of reducing the emergence of antibiotics resistance using anti-virulence and multi-targeting approaches.
Recent Publications
Contact
Gitai Lab
Department of Molecular Biology
Princeton University
355 Thomas Laboratory
Washington Road
Princeton, NJ 08544
p 609-258-9420
Faculty Assistant
Ellen Brindle-Clark
[email protected]
230 Thomas Laboratory
p 609-258-5419
f 609-258-6175
Lab Website
scholar.princeton.edu/gitailab

